A vision for extensibility to GPU & distributed support for SciPy, scikit-learn, scikit-image and beyond
Over the years, array computing in Python has evolved to support distributed arrays, GPU arrays, and other various kinds of arrays that work with specialized hardware, or carry additional metadata, or use different internal memory representations. The foundational library for array computing in the PyData ecosystem is NumPy. But NumPy alone is a CPU-only library - and a single-threaded one at that - and in a world where it's possible to get a GPU or a CPU with a large core count in the cloud cheaply or even for free in a matter of seconds, that may not seem enough. For the past couple of years, a lot of thought and effort has been spent on devising mechanisms to tackle this problem, and evolve the ecosystem in a gradual way towards a state where PyData libraries can run on a GPU, as well as in distributed mode across multiple GPUs.
We feel like a shared vision has emerged, in bits and pieces. In this post, we aim to articulate that vision and suggest a path to making it concrete, focusing on three libraries at the core of the PyData ecosystem: SciPy, scikit-learn and scikit-image. We are also happy to share that AMD has recognized the value of this vision, and is partnering with Quansight Labs to help make it a reality.
With the set of dispatch layers sketched in this post, array-consuming libraries will be able to use multiple kinds of arrays, without having to have a hard dependency on all of those array libraries. We'd like to have a unified design, using the same tools applicable across SciPy, scikit-learn, scikit-image and other libraries. In this situation, it's best to have a design discussion spanning multiple projects.
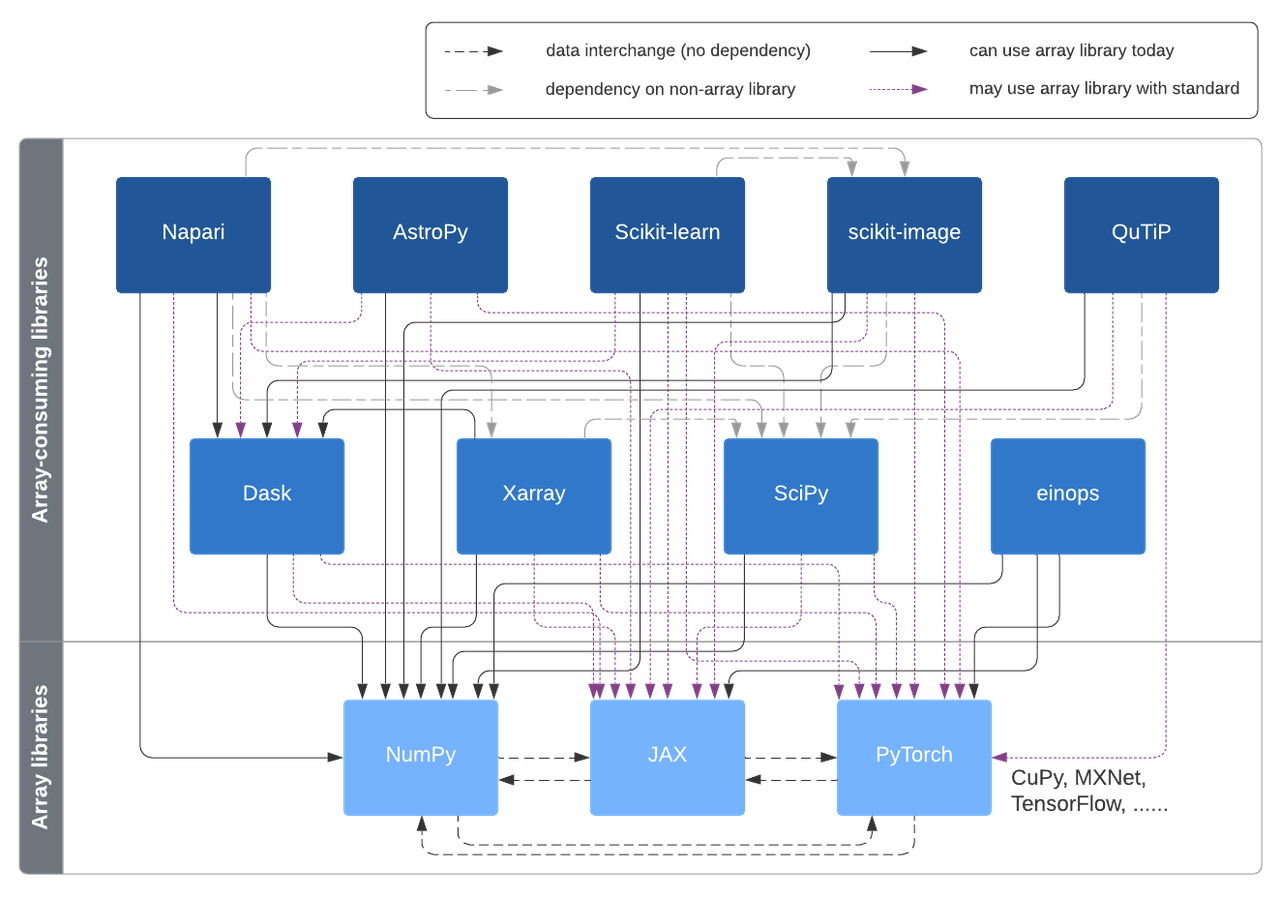
Possible interconnections enabled in this design, via the Array API standard and Uarray.
Today, SciPy, scikit-learn and scikit-image only work with NumPy arrays. SciPy has had "support for distributed and GPU arrays" on its roadmap for a couple of years. Parallel execution and GPU support were two of the top three priorities named by the 1000+ users in the 2020 NumPy user survey. The scikit-learn technical committee listed "evaluate interoperability with other types of arrays that are compatible with the NumPy API" as one of its 2020-2021 priorities. And scikit-image included exploring multi-threading and GPU acceleration in its (awarded) CZI EOSS grant proposal. Clearly there is a lot of interest in this topic.
Despite NumPy being a CPU-only library, it still can be used with alternative arrays due to its dispatch mechanism allowing users and library authors to write backend and hardware agnostic code using the NumPy API. When passing as input an alternative array (say from CuPy or Dask) to such generic code, the hardware-specific implementation is invoked instead of the internal to NumPy implementation. The NumPy API is a powerful tool for writing generic code and it is followed by several array libraries. Some libraries, like JAX and CuPy, had NumPy compatibility from the start, while other libraries, like PyTorch, are gradually moving towards it.
One of the problems of using the NumPy API as a reference standard is that it
wasn't designed with different types of hardware in mind and has a number of
inconsistencies that make it difficult for other libraries to reimplement its
API (in addition to that API being very large and a little ill-defined).
The Array API standard
solves this problem by standardizing functionality across most array libraries.
NumPy and CuPy have already adopted the Array API standard; it can be accessed
via the __array_namespace__ mechanism in NumPy >=1.22.0 and CuPy >=10.0.0
(both to be released in the coming month).
The common architecture of SciPy, scikit-learn, and scikit-image is that they use NumPy for array operations, and critical-to-performance parts are written using compiled code extensions. Unfortunately, compiled extensions rely on NumPy's internal memory representation and are restricted to CPU computations. For these reasons, Python code with compiled extensions is not compatible with NumPy's dispatching capabilities; it would not know how to deal with a GPU array. It's also very challenging to rewrite the libraries to use pure NumPy without sacrificing performance. Therefore we need a dispatching mechanism (similar to NumPy's) for the API of modules in SciPy and other projects that rely on compiled extensions.
Let's imagine some SciPy module that equally depends on compiled code and NumPy
functions. Removing all forced conversions to NumPy arrays inside the module
would open the "Dispatcher" path, but only a small part of SciPy
functionality would work for generic arrays. In the diagram, the NumPy
Dispatcher is an abstract mechanism, concretely it could be an
__array_function/ufunc__-based one for older versions of NumPy or
__array_namespace__ - for NumPy 1.22+.
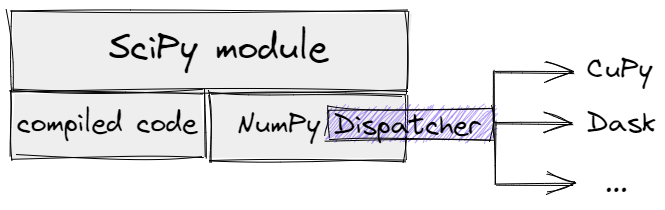
A SciPy module with NumPy "Dispatcher".
For SciPy and similar libraries, we need to follow NumPy's example and add our own dispatching layer for the API to allow users to develop generic code that works with arrays from different libraries. Then it would be possible to directly reroute to specific array implementations bypassing compiled code paths.

A SciPy module with its own "Dispatcher".
A careful reader may note that there's a gray area left: what if some SciPy function or submodule is mostly pure Python + NumPy, but relies on a small amount of compiled code (which may be private API) for performance? It may then not be completely clear what should be done. In some cases it may make sense to make that private API public and dispatch on it, in other cases dispatching should happen at the top-level - and perhaps there are better solutions to allow reuse of the pure Python code.
Goals, wishes, and constraints
The main goal is to separate the interface from the implementation and let the dispatching system help users and library authors write generic code that works with different kinds of arrays.
The new dispatching system implemented for SciPy and scikits should:
- add the possibility to support alternative array libraries, not just NumPy
- provide a way to select a different backend for the same array type (examples:
scipy.fft+ alternative CPU FFT libraries like pyFFTW, or scikit-learn + an Intel optimized implementation) - support "array duck types", i.e. custom array objects that are compatible
with a particular array library via (for example)
__array_function__or__torch_function__. - be able to extend function signatures in a backward-compatible way on the base library side (all alternative implementations should not break after the change)
- have a performance overhead that is as low as possible
- have a reasonably low maintenance overhead (e.g., scikit-learn has explicitly stated that they cannot afford GPU-specific code in their codebase).
- ideally be adoptable also for libraries that now default to another array library than NumPy (e.g., PyTorch, JAX, TensorFlow, CuPy, and Dask - they all have growing and thriving ecosystem of their own).
When designing API override systems, there are many possible design choices. Three major axes of design decisions in the context of the NumPy API overrides are outlined in the appendix of NumPy Enhancement Proposal (NEP) 37; they are also applicable outside of NumPy:
- Opt-in vs. opt-out for users
- Explicit vs. implicit choice of implementation
- Local vs. non-local vs. global control.
A concrete design proposal
In the figure below, we sketch a design for implementing this backend & dispatch system in SciPy, scikit-learn, scikit-image and other core projects which now rely only on NumPy. It uses CuPy and Dask as examples, with modules that exist today. The design is generic though, and will also work for other libraries with compatible APIs like PyTorch and JAX once they support this system.
We need two dispatching mechanisms: one for the modules with compiled code
portions, and one for pure Python code.
The first dispatching mechanism is based on the uarray
project, which is a backend dispatcher that allows us to
choose the relevant function implementation at runtime. The second dispatching
mechanism is based on the __array_namespace__ method from the Array API
standard, which is a method that allows us to get the Array API implementation
module specific to CuPy (or any other array library) at runtime.
__array_namespace__ will be available in NumPy 1.22 and CuPy v10.0.
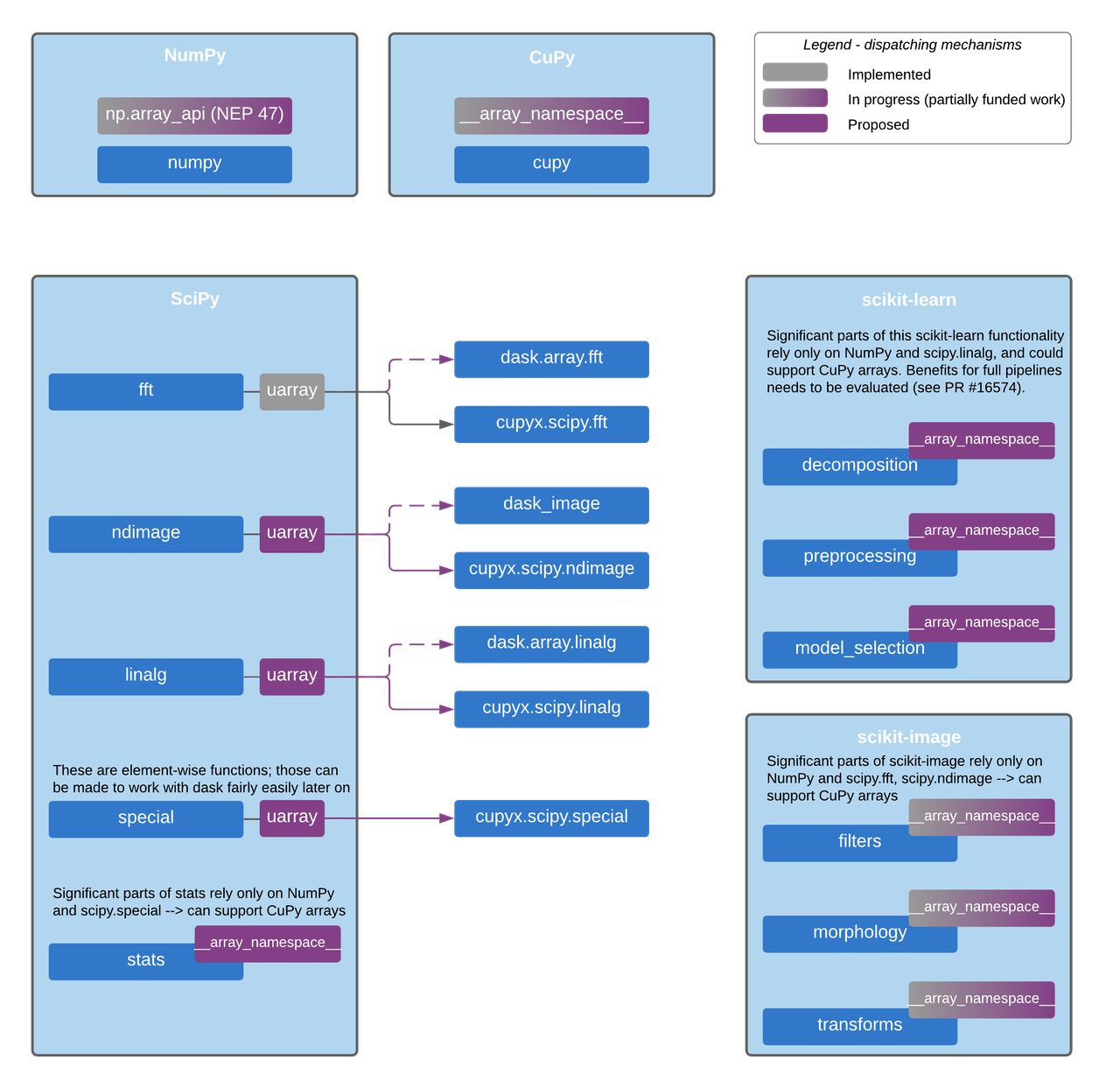
Proposed dispatch mechanism layers for enabling CuPy and Dask support.
This will also support any other array library with the same Array API standard support and uarray backends.
One of the key questions to address in this design is whether the behavior should be opt-int or opt-out for users. And if it's enabled by default by a library (and hence opt-out for users, or not even giving users a choice), is it the library which implements the n-D array object that controls this enabling or is it individual array-consuming libraries?
We think that the individual array-consuming libraries should control the
dispatch. And the dispatching itself should be turned off by default for
backwards-compatibility. The same switch should control enabling uarray
dispatching and use of __array_namespace__ simultaneously to have consistent
behavior.
Using the Array API standard
The Array API standard is a specification of functionality that array libraries need to implement. There is a dedicated test suite to verify the compliance of specific implementations with the standard, so it's easy to check all libraries adopting the standard have a unified behavior. NEP 47 has a couple of examples on usage of the Array API namespace.
__array_namespace__ is a central piece of the Array API dispatch method. It is
a method of an array object that returns an object that has all the array API
functions on it. In the case of NumPy calling __array_namespace__() would
return the numpy.array_api module. The __array_namespace__ method attached to
the array object enables downstream libraries to consume multiple kinds of
arrays, without having to have a hard dependency on any of those array
libraries. The pattern to support multiple array libraries inside, say, a SciPy
function is intended to be similar to the following code snippet:
def somefunc(x, y):
# Retrieves standard namespace.
# Raises if x and y have different namespaces.
xp = get_namespace(x, y)
out = xp.mean(x, axis=0) + 2*xp.std(y, axis=0)
return out
Notice that to make the above code valid, we don't need to import the package
that implements the mean and std functions explicitly; the required module
is stored in the x and y variables.
For several concrete use cases of the Array API standard, check out use case
descriptions in the standard itself.
Anirudh Dagar also wrote a great
demo
adapting a LIGO gravitational waves tutorial and showcasing what it would take to
support PyTorch tensors for a subset of the SciPy API using __array_namespace__.
In the accompanying blog post, titled Array Libraries
Interoperability,
Anirudh explores array interoperability methods: __array_function__
(NEP 18), __array_namespace__ (NEP 47) and uarray.
PyData dispatching system
In the future, we'd like to pass alternative arrays to SciPy and scikits
functions and get the results without transforming internally to NumPy arrays.
Instead, it would automatically handle dispatching to the appropriate
implementation, for example, cupyx.scipy for GPU-accelerated SciPy functions,
scikit-learn-intelex for Intel optimized version of scikit-learn, or cuCIM for
GPU-accelerated scikit-image. Usually in statically typed languages function
overloading is used to implement a function with the same name but arguments of
different types. How can we implement something similar to function overloading
but in Python? Well, we could implement one central function checking the types
of the arguments using isinstance(), or issubclass() and calling appropriate
implementations based on these checks. This approach doesn't scale well with the
number of inputs of different types (see
here
for a concrete example) and is not extendable by third-party
implementers without modifying the source code. A second approach is monkey
patching the original library (this is what scikit-learn-intelex does now
for example); that is a fragile method that can break at any time when the
library being patched makes changes to its internals or API.
Another approach is to use
multiple dispatch, where each function has multiple variants that are chosen
based on dynamically determined types. Unfortunately, multiple dispatch
is not built-in for Python, but there's a simpler version - single
dispatch. For a demo, let's begin with a tool from the Python standard library:
functools.singledispatch.
from functools import singledispatch
import scipy.ndimage as _scipy_ndimage
@singledispatch
def laplace(input, output=None, mode="reflect", cval=0.0):
return _scipy_ndimage.laplace(input, output=output, mode=mode, cval=cval)
# export `laplace` as `scipy.ndimage.laplace`
# In CuPy codebase
# from scipy.ndimage import laplace
import cupyx.scipy.ndimage as _cupyx_ndimage
@laplace.register(cupy.ndarray)
def cupy_laplace(input, output=None, mode="reflect", cval=0.0):
return _cupyx_ndimage.laplace(input, output=output, mode=mode, cval=cval)
# run registration code when CuPy is imported
We begin by defining the base laplace function, it is a fallback
implementation that is going to be called for non-registered types of input.
Next laplace.register is used to connect the implementation that is specific
to cupy.ndarray with the base laplace function. We get a function in the
scipy.ndimage module that is now compatible with CuPy arrays. Similarly, it
can be extended to work with Dask arrays.
from scipy.ndimage import laplace
import numpy as np
import cupy as cp
numpy_img = np.random.normal(size=(256, 256))
cupy_img = cp.array(numpy_img)
print(type(laplace(numpy_img)))
# <class 'numpy.ndarray'>
print(type(laplace(cupy_img)))
# <class 'cupy._core.core.ndarray'>
The above single-dispatch example illustrates the dispatching principle, but is too simple for our purposes - so let's look at multiple dispatch methods. The Python stdlib doesn't provide tools for multiple dispatch, however there are several dedicated libraries:
- multimethod - pure Python implementation of multiple dispatch with caching of argument types
- Plum - implementation of multiple dispatch that follows the ideas from Julia
- multipledispatch - similar to Plum, only with slightly fewer features and no longer developed; vendored in SymPy
-
uarray is a generic
backend/multiple dispatch library similar to NumPy's dispatch functionality
except that the implementation doesn't need to be baked into the array type;
used in SciPy's
fftmodule.
The first three options are very similar in functionality and usability. But the last option (uarray), in addition to multiple dispatch, offers granular control using context managers and provides a way to switch backends for the same array type. It is also the only library that doesn't solely rely on a class hierarchy/relationship, so can support "array duck types".
It's also possible to make the uarray dispatcher work without a context
manager making the mechanism implicit and similar to other dispatch libraries.
Whether we should encourage the implicit registration of backends or not is an
open design issue and there's previous discussion on the topic in
SciPy #14266. Hameer Abbasi, one of the
uarray authors, wrote
a blog post
about the motivation for uarray and how it compares to NumPy's
__array_function__ dispatch mechanism.
In 2019, Peter Bell added support for backend switching to the scipy.fft module
(PR #10383) using uarray. Now
it's possible to tell SciPy to use, for example, the CuPy backend, for
computing FFT when CuPy's array is passed to functions from scipy.fft.
Next steps
This post is the invitation to discuss the idea on the Scientific Python's forum, a recent website for better coordinating the Scientific Python ecosystem. The forum is a place for discussing the general idea of array interoperability, and it's a good place to give feedback on this proposed design.
One important aspect of the discussion is to discuss the design choices for the dispatch API. We particularly need to come to a conclusion on the topic of "opt-in vs opt-out". Once there's a consensus on the design with maintainers of SciPy, scikit-learn, scikit-image and the wider community, we can move forward with the implementation.
There are a couple of PRs that are already in the queue. The first one is adding
uarray-based backend switching to scipy.ndimage
(PR #14356) and the second one is adding
it to scipy.linalg.decomp (PR #14407).
After these PRs are merged, we will explore using the array API standard in
SciPy and scikit-learn with CuPy v10.0.
There's a lot of prior community effort to make array interoperability in the PyData ecosystem better. This is an exciting project, and we're looking forward to making progress on this front!
Acknowledgment
Adding dispatching support will take significant effort to implement across different projects. Fortunately, AMD recognized this as a problem worth solving and is willing to help to fix it by funding a team at Quansight Labs to do the initial heavy lifting.
AMD's vision for the developer community is open, portable tools that enable the same source code to run on different hardware platforms. ROCm is an example of this, supporting both OpenMP and HIP on multiple architectures. "AMD and Quansight are aligned in this direction. We believe that by working together and working with the Python community, we can achieve an open, portable solution for all Python developers," says Terry Deem, AMD Product Manager for ROCm.
Comments