Improving the interpolation and signal processing capabilities of CuPy
Published November 23, 2023
andfoy
Edgar Margffoy
CuPy, the NumPy/SciPy-compatible array library for GPU-accelerated computing, is getting better every release. In this post, we'll take a deep dive into the new interpolation and signal processing capabilities that our team has been adding to CuPy.
For those who are not familiar with the project, CuPy has several unique features making it an excellent choice for numerical computation. First, CuPy is a drop-in replacement for NumPy and SciPy and used to perform numerical computation on GPUs, thus allowing developers to create, manipulate, and operate on larger n-dimensional arrays faster than they can on CPUs. As a drop-in replacement, CuPy helps users without knowledge of GPU programming accelerate their existing pipelines, while also accommodating more experienced programmers who want to write low-level GPU routines directly in Python. Second, CuPy conveniently integrates with popular Deep Learning frameworks, such as PyTorch and TensorFlow, and allows developers to efficiently deploy new operators, modules, and functions. And lastly, CuPy is flexible with respect to the hardware used, allowing kernel execution not just on NVIDIA GPUs (via CUDA), but also on AMD GPUs. Support for AMD GPUs is thanks to the HIP integration effort pushed during the past year, thus making CuPy a universal, vendor-free library that can be used regardless of the GPU accelerator available.
Over the past few years, CuPy's compatibility with NumPy has improved to the point that most commonplace and popular APIs are now available, including n-dimensional array creation, operation, and manipulation, linear algebra, fast Fourier transforms (FFTs), and statistics. Given the great progress in replicating the NumPy API on GPU, the next step was to port the extensive SciPy API to support GPUs, as well. This has led to several improvements, such as sparse array support and spatial computation.
Porting SciPy APIs is a significant undertaking. Until recently, there were
several modules, such as interpolation and signal processing, that were missing
or had a partial API coverage and are critical in scientific
applications like image and audio processing, weather, radar and spectral analysis,
as well as computational chemistry and sensor design. Given this interest, a
joint CZI grant (EOSS Cycle 5) was proposed between
Preferred Networks and Quansight Labs in order
to reduce the gap between SciPy and CuPy and to specifically port most of the
APIs available in the scipy.interpolate and scipy.signal namespaces.
During the first year of the grant work, a team of developers at Quansight Labs have teamed up with CuPy maintainers at Preferred Networks to increase the coverage of interpolation and signal processing subpackages, where, as of CuPy pre-release 13.0.0b1, 18 interpolation and more than 100 signal APIs are now available, including basis splines, regular grid interpolation, and piecewise polynomials for the interpolation module and IIR linear filtering, filter design, linear time invariant systems, and peak finding for the signal processing subpackage.
As expected, all the ported APIs afford a significant performance increase
relative to the SciPy CPU reference implementations by exploiting the parallel
nature of GPU kernels (whereas CPU implementations are sequential).
For example, the new CuPy BSpline implementation has a nearly 10x performance
improvement when compared to its corresponding implementation in SciPy,
and PPoly has a 32x performance improvement, representing a huge enhancement
over CPU implementations.
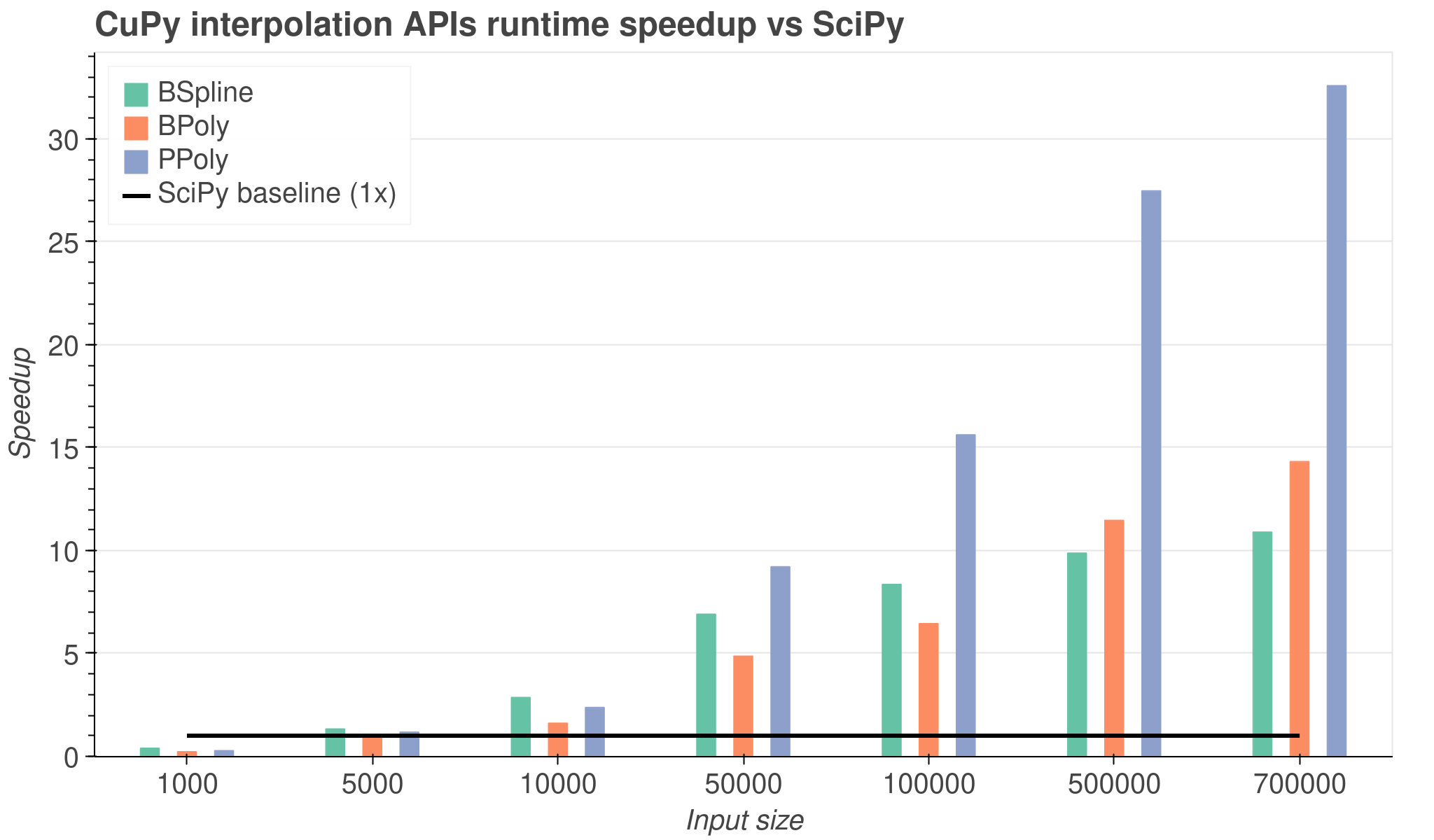
Relative speedup of CuPy’s BSpline, PPoly and BPoly interpolation classes relative to SciPy for various input sizes.
For those APIs which could not be readily ported from CPU to GPU, new implementations were derived from scientific papers, including the new IIR linear filtering routines, which represent the first time IIR filters can be applied on GPUs from Python. As part of this process, some enhancements and simplifications were also backported to SciPy, such as a major refactor of symmetric IIR filters to leverage IIR linear filtering and second order sections rather than dedicated routines which contained duplicate code.
The following example demonstrates the new capabilities of CuPy in filtering multiple signals in parallel using a GPU. For illustrative purposes, we begin by defining four different signals.
import cupy as cpfrom cupyx.scipy import signalimport matplotlib.pyplot as pltrng = cp.random.default_rng(seed=1234)t = cp.linspace(-1, 1, 201)t = cp.broadcast_to(t, (4, 201)).copy()phase = 10 * rng.random((4, 1)) - 5# Define 4 random signals composed of a main component with quadratic time# input and two sinusoids with 1.25Hz and 3.85Hz frequencies, plus a DC component.main_x = cp.sin(2 * cp.pi * 0.75 * t * (1 - t) + 2.1 + phase)
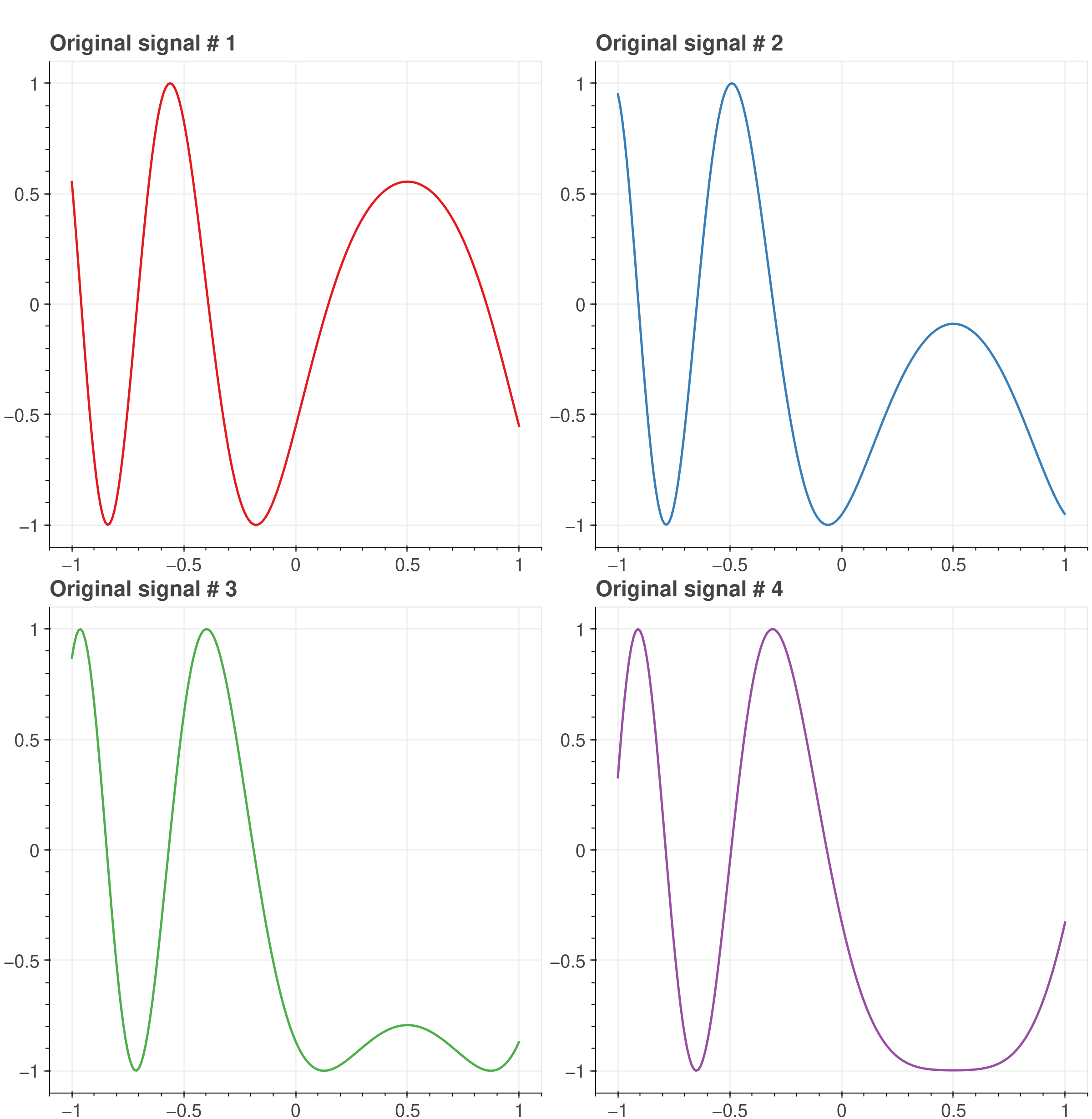
Four random signals composed of a main component with quadratic time input and two sinusoids with 1.25Hz and 3.85Hz frequencies, plus a DC component.
Next, let’s add some noise to the example signals.
# Introduce linear noise into the original signal.x = (main_x + 0.1 * cp.sin( 2 *cp.pi * 1.25 * t + 1) + 0.18 * cp.cos(2 * cp.pi * 3.85 * t))xn = x + rng.standard_normal(t.shape) * 0.08# Make sure that input is float32, since it yields a better performancexn = xn.astype(cp.float32)
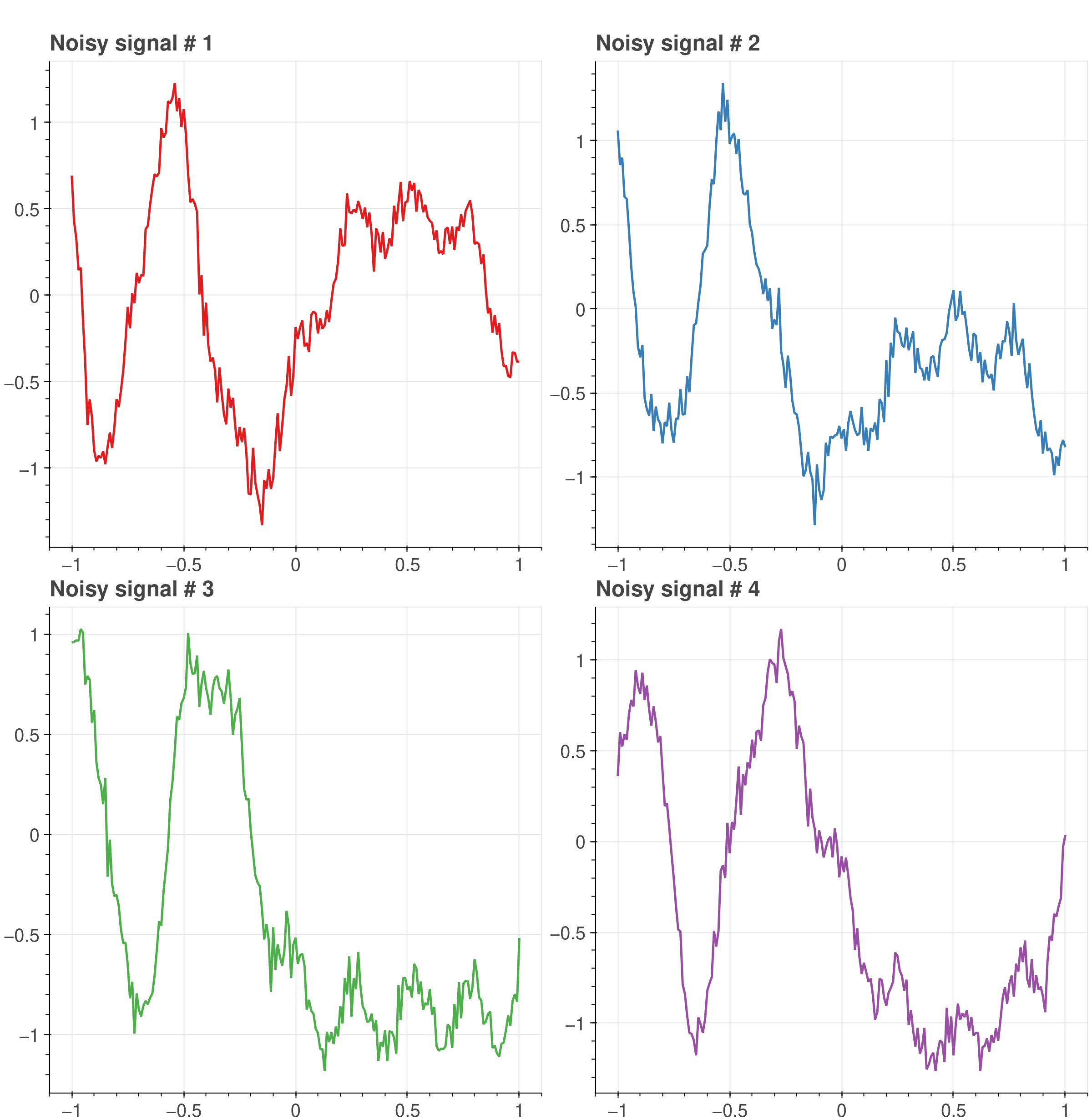
The four random input signals from the previous figure with added Gaussian noise.
Now that we’ve generated our signals, time for the fun part! Let’s do some filtering!
In doing so, we’ll extract the noise from each signal using the new signal
parallel-processing APIs available in CuPy, namely lfilter and filtfilt.
For purposes of comparison, we’ll compare applying lfilter once, applying
lfilter twice, and filtering in both forward and reverse directions
using filtfilt.
# Create a third-order, Butterworth low-pass filter with cutoff at 0.05.b, a = signal.butter(3, 0.05)b = b.astype(cp.float32)a = a.astype(cp.float32)# Compute the steady-state response of the filter.zi = signal.lfilter_zi(b, a)zi = zi.reshape(1, -1)zi = zi.astype(cp.float32)# Filter out the 1.25Hz and 3.85Hz components and remove the noise on all signals in parallel.z, _ = signal.lfilter(b, a, xn, zi=zi * xn[:, 0, None])# Apply the filter again.z2, _ = signal.lfilter(b, a, z, zi=zi*z[:, 0, None])# Use filtfilt on all signals in parallel.y = signal.filtfilt(b, a, xn)
A plot comparing filtering results is displayed below. By visual inspection
(and rather cursory analysis!), we can conclude that filtfilt does a better
job at recovering the original signals.
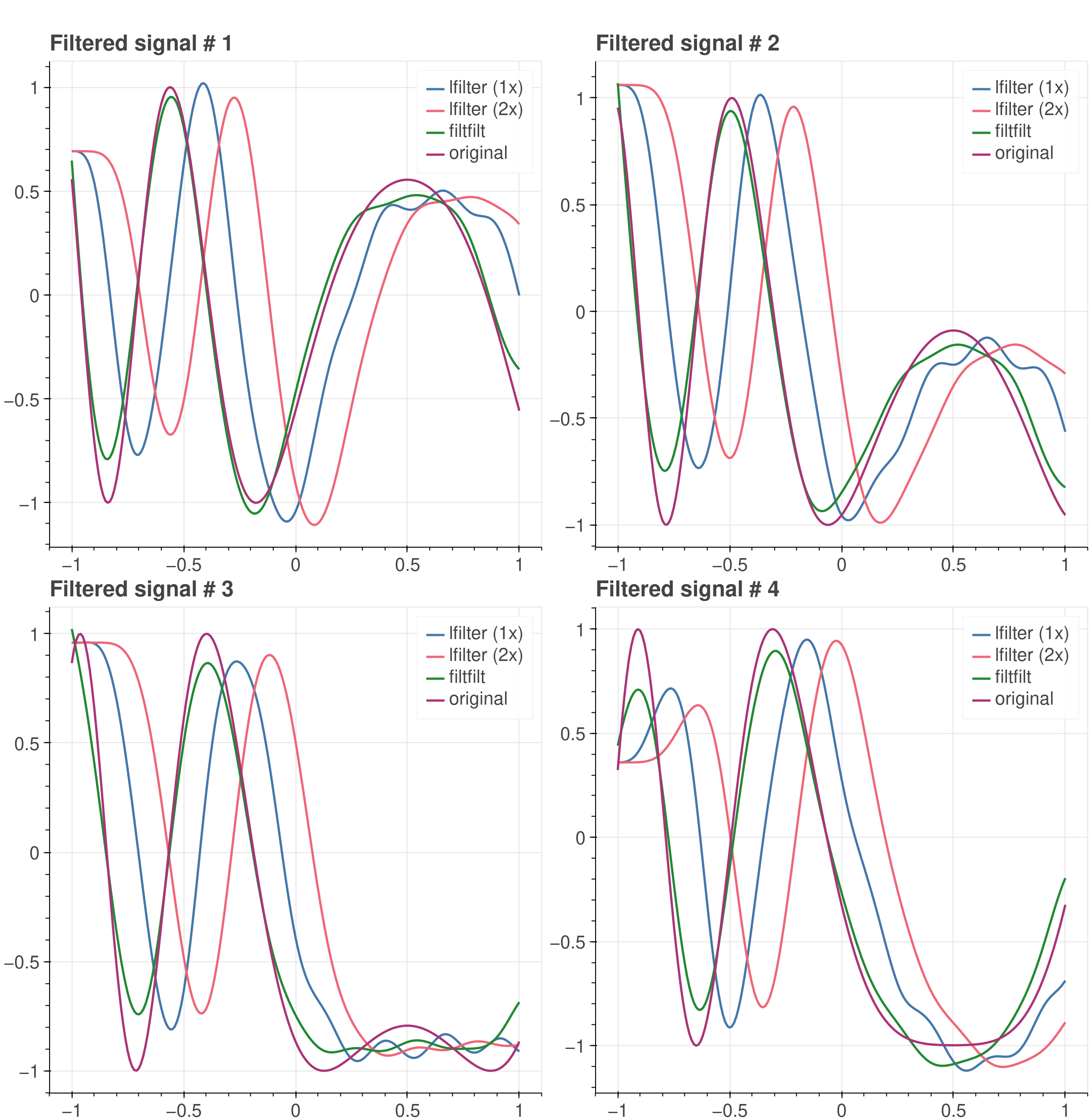
Filtering results using the new lfilter and filtfilt CuPy APIs. Each result is compared against the original, non-noisy signal.
As part of this effort, we made several other improvements to other CuPy modules,
such as adding special functions for elliptic filters, implementing the
matrix exponential in the cupyx.scipy.linalg namespace, and implementing
KD-Trees in cupyx.scipy.spatial.
This work would not have been possible without the help of the
cuSignal team at the NVIDIA RAPIDS
initiative, who agreed to migrate the implementations of over 36 APIs from their project to
CuPy, thus unifying the GPU signal processing efforts currently available
in the ecosystem. As part of this migration, cuSignal will be deprecated and
replaced by the new cupyx.scipy.signal module calls. The calls will be
interchangeable by simply replacing the cusignal imports with imports from
cupyx.scipy.signal.
As the grant enters its second year, we plan to finish the overall interoperability of the interpolation module, which first requires a significant refactor of several FORTRAN routines for spline smoothing into GPU kernels and, second, implementing computational geometry routines to compute convex hulls and Delaunay triangulations, which are used for interpolating multi-dimensional unstructured data. This work will further enhance overall CuPy-SciPy compatibility.
These new APIs will open a sea of possibilities in terms of acceleration and batch processing of data, and we hope they will be useful to the CuPy community. Right now, these additions are being pre-released as part of the beta releases of the upcoming CuPy v13.0. We invite you to keep an eye on our tracking issues https://github.com/cupy/cupy/issues/7186 and https://github.com/cupy/cupy/issues/7403, where you can find more information on and discussions about implementations, as well as the current progress on the remaining functionality of CuPy’s interpolation and signal modules. Please feel free to chime in and contribute to any discussion or improvement to the project!
We would like to thank the Chan Zuckerberg Initiative (CZI), Preferred Networks, and Quansight Labs for the resources used to plan and execute the current grant. We would also like to thank NVIDIA and the RAPIDS team for their help and willingness to migrate cuSignal into CuPy as part of this work.
In particular, we would like to thank the following individuals on the CuPy team at Preferred Networks: Masayuki Takagi (@takagi), Emilio Castillo (@emcastillo), Akifumi Imanishi (@asi1024) and Kenichi Maehashi (@kmaehashi). Shout out to the Quansight Labs team: Evgeni Burovski (@ev-br), Edgar Margffoy (@andfoy), and Ralf Gommers (@rgommers). And we would like to thank Adam Thompson (@awthomp) and Matthew Nicely (@mnicely) on the cuSignal team for their great effort in making CuPy a complete GPU replacement for NumPy and SciPy.
Finally, we thankfully acknowledge the community contributions to this effort.
Specifically, we thank Michael Zhang (@ideasrule)
for his porting the RegularGridInterpolator code from SciPy and
Leo Fang (@leofang) of NVIDIA for multiple
fruitful discussions, as well as other community members for their
feedback and suggestions.